Mike Livermore – CSI 773 – Week 8 Assignment –Maps and Smoothes – November 2010
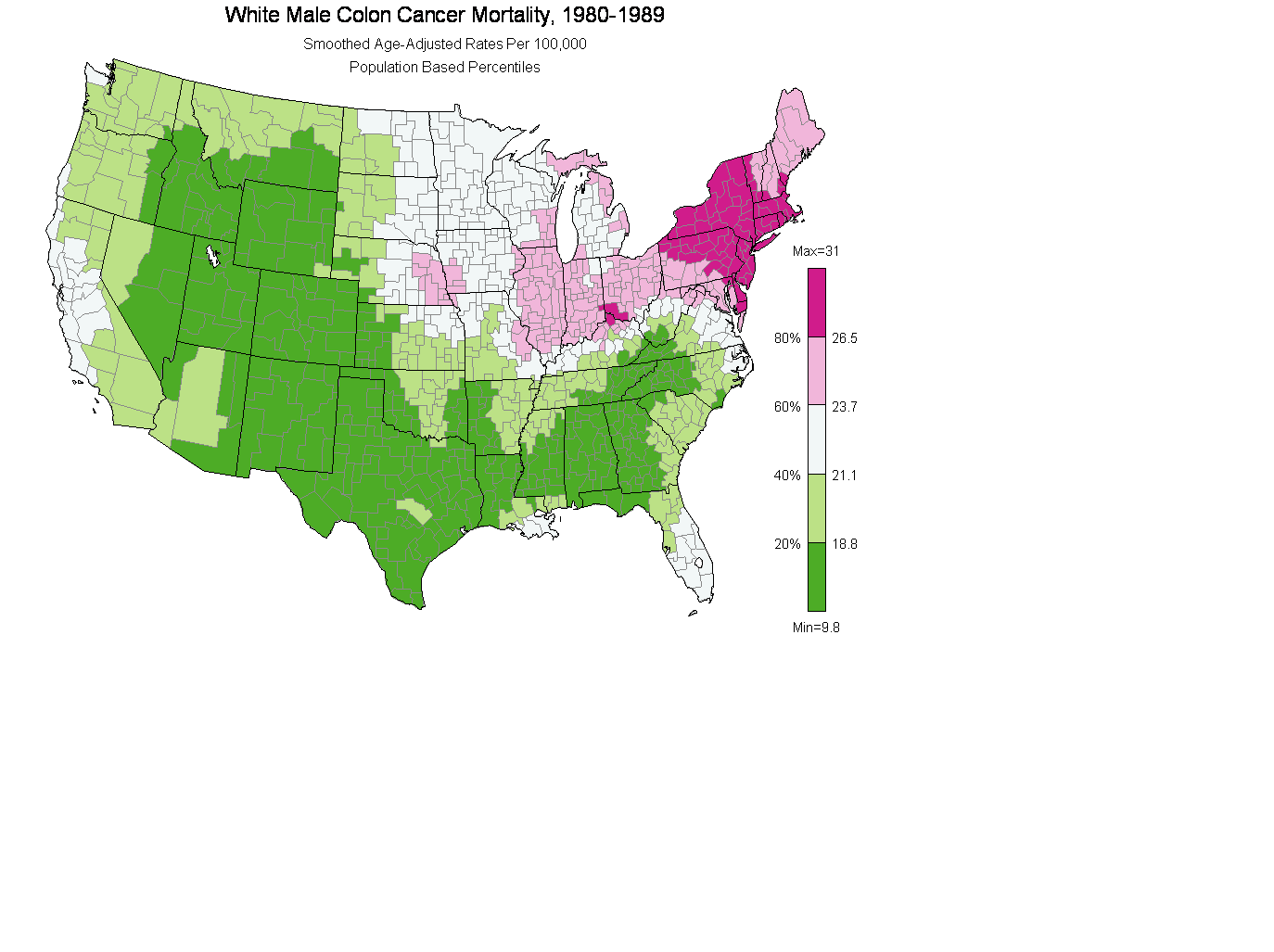
(1) Plot 4.2 Region centroids smoothed map with Green Violet palette
(2) Model 3 compared with Model 2
model3 = loess(rate ~ a.long*a.lat,
span=.20,
degree=2,
surface='direct',
normalize=FALSE,
weights=pop/(10^5*rate))
summary(model3)
Call:
loess(formula = rate ~ a.long * a.lat, weights = pop/(10^5 *
rate), span = 0.2, degree = 2, normalize = FALSE, surface = "direct")
Number of Observations: 802
Equivalent Number of Parameters: 35.89
Residual Standard Error: 1.485
Trace of smoother matrix: 42.89
Control settings:
normalize: FALSE
span : 0.2
degree : 2
family : gaussian
surface : direct
it = 2
for (i in 1:it){
model3 = loess(rate ~ a.long*a.lat,
span=.20,
degree=2,
surface='direct',
normalize=FALSE,
weights=pop/(10^5*fitted(model3)))
}
summary(model3)
Call:
loess(formula = rate ~ a.long * a.lat, weights = pop/(10^5 *
fitted(model3)), span = 0.2, degree = 2, normalize = FALSE,
surface = "direct")
Number of Observations: 802
Equivalent Number of Parameters: 35.93
Residual Standard Error: 1.426
Trace of smoother matrix: 42.93
Control settings:
normalize: FALSE
span : 0.2
degree : 2
family : gaussian
surface : direct
anova(model1, model3)
Model 1: loess(formula = rate ~ a.long * a.lat, weights = pop/(10^5 * fitted(model1)), span = 0.15, degree = 2, normalize = FALSE, surface = "direct")
Model 2: loess(formula = rate ~ a.long * a.lat, weights = pop/(10^5 * fitted(model3)), span = 0.2, degree = 2, normalize = FALSE, surface = "direct")
Analysis of Variance: denominator df 742.87
ENP RSS F-value Pr(>F)
[1,] 46.49 1444.5
[2,] 35.93 1528.4 2.9349 0.0002791 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
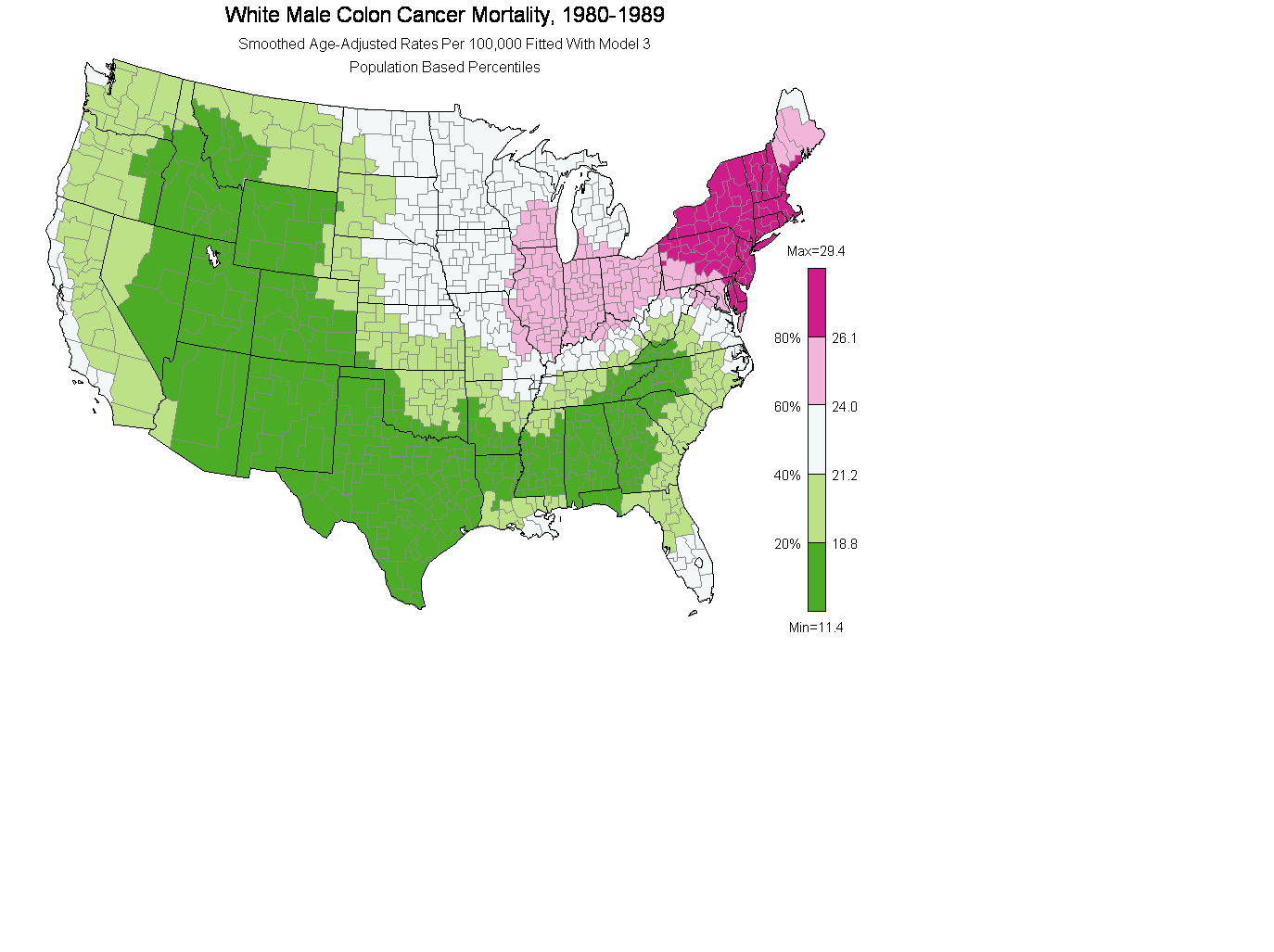
(3) Smoothed Plot 4.2 fitted with model 3:
hsaSmooth = fitted(model3)
rateClass = classifyRates(hsaSmooth,pop,percents)
polygonClass =classifyPolygons(hsaPolyId,rateId,rateClass$class)
hsaSetup()
polygon(hsaPoly,col=currentPalette[polygonClass],density=-1,border=F)
polygon(hsaPoly,col="#898989",density=0,lwd=1)
polygon(usGrid90States,density=0,lwd=1,col="black") # "#606060"
mtext('White Male Colon Cancer Mortality, 1980-1989',side=3,line=.7,cex=1.4,adj=.5)
mtext('Smoothed Age-Adjusted Rates Per 100,000 Fitted With Model 3', side=3,line=-.8,cex=1.0,adj=.5)
mtext('Population Based Percentiles', side=3,line=-2.1,cex=1.0,adj=.5)
albers.legend(percents,rateClass$breaks,palette=currentPalette,brk.rnd=1)
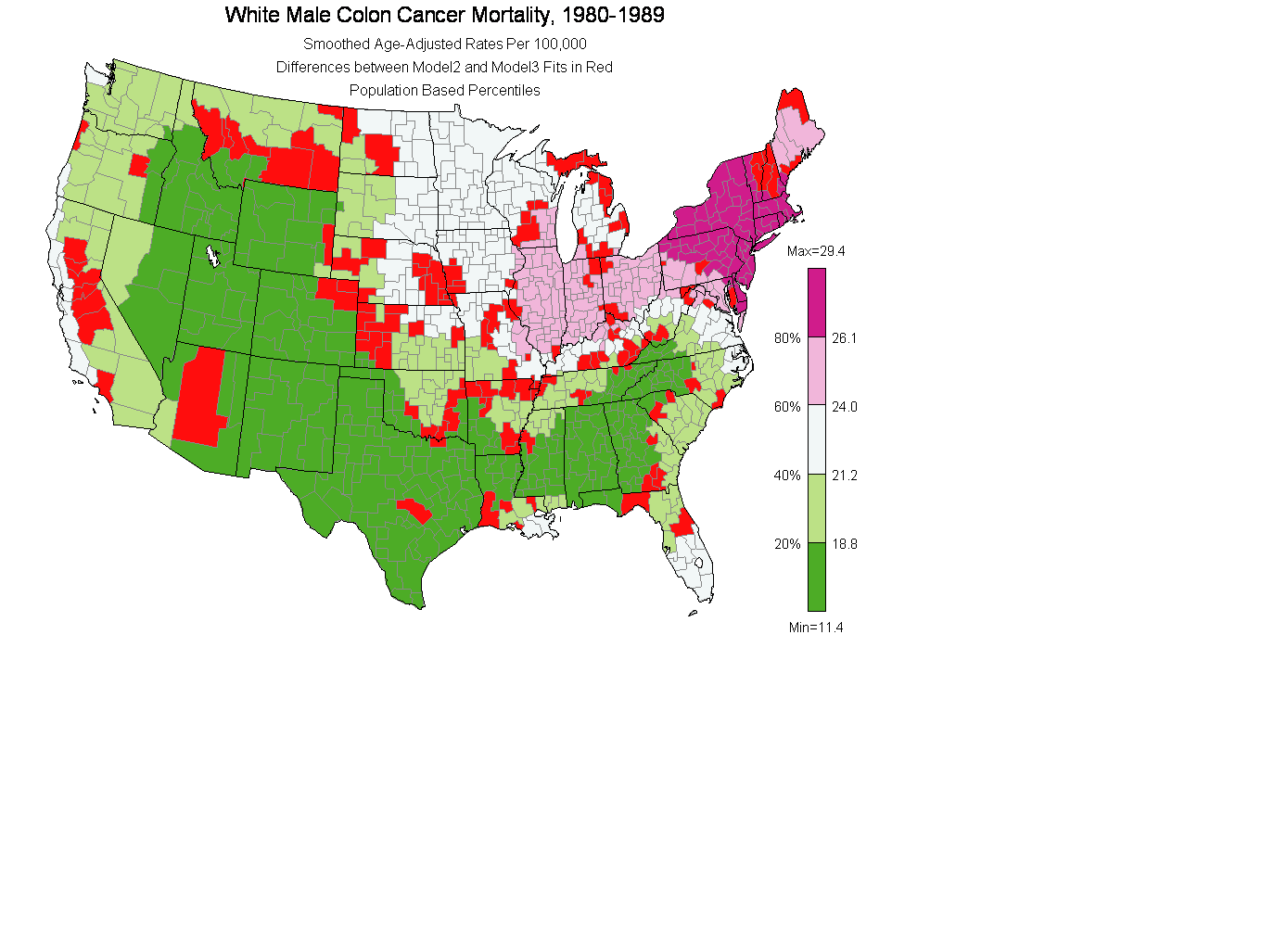
(4) Show Differences between 4.2 model 2 and model 3
currentPalette[6]=blueRed5[5]
hsaSmooth2 = fitted(model2)
rateClass2 = classifyRates(hsaSmooth2,pop,percents)
polygonClass2 =classifyPolygons(hsaPolyId,rateId,rateClass2$class)
replaceDiff = function(list1, list2, newValue) {
listRet = list1
for(index in 1:length(list1)) {
if (list1[index] != list2[index]) listRet[index] = newValue
}
return (listRet)
}
polygonClass = replaceDiff(polygonClass, polygonClass2, 6)
hsaSetup()
polygon(hsaPoly,col=currentPalette[polygonClass],density=-1,border=F)
polygon(hsaPoly,col="#898989",density=0,lwd=1)
polygon(usGrid90States,density=0,lwd=1,col="black") # "#606060"
mtext('White Male Colon Cancer Mortality, 1980-1989',side=3,line=.7,cex=1.4,adj=.5)
mtext('Smoothed Age-Adjusted Rates Per 100,000', side=3,line=-.8,cex=1.0,adj=.5)
mtext('Differences between Model2 and Model3 Fits in Red', side=3,line=-2.1,cex=1.0,adj=.5)
mtext('Population Based Percentiles', side=3,line=-3.4,cex=1.0,adj=.5)
albers.legend(percents,rateClass$breaks,palette=currentPalette,brk.rnd=1)